A novel mutation in CELSR1 is associated with hereditary lymphedema
Vascular Cell. 2016;
Received: 8 October 2015 | Accepted: 29 January 2016 | Published: 5 February 2016
Vascular Cell ISSN: 2045-824X
Abstract
Background
Biological evidence reported in the literature supports the role of CELSR1 as being essential for valvular function in murine lymphatics. Yet thus far, there have been no variants in
Case Presentation
In this report, a rare early inactivating mutation in
Conclusion
Keywords
Primary lymphedema Whole exome sequencing Near-infrared fluorescence imaging CELSR1 Planar polarityBackground
Lymphedema (LE) is a condition caused by a defective lymphatic system where an excess of fluid accumulates and generates chronic swelling of tissues. Primary LE is a rare genetic disorder that can develop at birth (congenital), at puberty (praecox), or after 35 years of age (tarda) [1].
In this report, we present the unusual lymphatic phenotype obtained by near-infrared fluorescence lymphatic imaging (NIRFLI) of a woman diagnosed non-syndromic, lower extremity LE and compare it with that of her asymptomatic mother. From clinical diagnoses and whole exome sequencing (WES) of family members across three generations, we found no gene variants known to be responsible for LE, but instead found a rare, inactivating mutation in
Case Presentation
Family case report
A 62 yo female previously diagnosed with primary, lower extremity LE (bilateral, Stage II by International Society of Lymphology staging system) and her 84 yo asymptomatic mother enrolled in an ongoing, institutional review board (IRB) and FDA-approved (IND# 102,827) clinical study of lymphatic disorders (Clinical Trials No. NCT00833599: “Imaging lymphatic function in normal subjects and in persons with lymphatic disorders” www.clinicaltrials.gov). The index case self-reported onset of lower extremity swelling at age 10, with diagnosis of LE at age 39 at which time she began compression treatment. In addition, she was also previously diagnosed with type 2 diabetes, high cholesterol, and high blood pressure. Following consent, a clinical examination was first performed followed by near-infrared fluorescence lymphatic imaging (NIRFLI) [5] to assess lymphatic anatomy and function in the legs. Blood was collected for WES of DNA.
Upon questioning, we further found a family history of lower extremity LE and traveled to the family’s hometown to consent, conduct standard clinical examinations (including measurements of leg volumes and assessment of Stemmer’s sign), and to collect blood from family members spanning three generations. We did not perform NIRFLI on these family members. Other than stage I and II bilateral lower extremity, non-syndromic LE, we found no remarkable evidence of disease among the family members.
Imaging studies
For lymphatic imaging of the daughter and mother, doses of 25 μg of indocyanine green (ICG) in 0.1 cc saline were administered bilaterally in the dorsum of the feet, medial ankles, the medial and lateral calves, and the thighs for a total dose of 300 μg. Images were acquired with exposure times of 200 ms and sequential images were compiled to assess for the presence of active lymphatic propulsion/movement of ICG-laden lymph from the injection sites proximally towards the inguinal lymph node basins. As shown by NIRFLI in Fig. 1b, the affected daughter had extensive areas of dermal backflow in her lower legs, as commonly observed in LE subjects, but possessed a unique, uncommon flow pattern of lymph movement that appeared in “sheets” rather than within distinct vessel lumens (Additional file 1: Video 1). While the proband’s mother primarily had well-defined and linear lymphatics (Fig. 1b) as typically seen in healthy subjects [6, 7], she nonetheless presented with smaller areas of dermal backflow and tortuous lymphatics as shown in the insets of Fig. 1b. Active lymphatic propulsion of ICG-laden lymph was observed in the well-defined lymphatic vessels in the mother (Additional file 2: Video 2), in contrast to the limited lymphatic movement observed in the daughter.
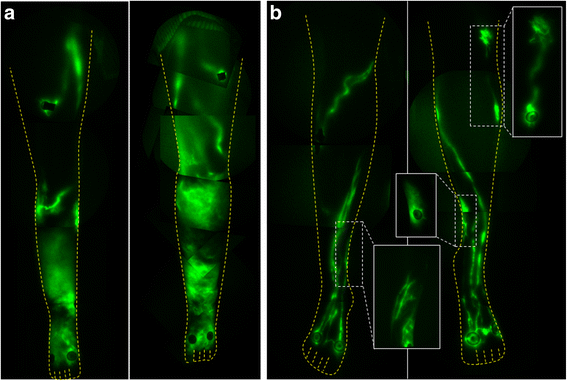
Fig. 1
Fig. 1 caption
Montages of NIRFLI of the lower extremities of (a) the proband and (b) her mother. While the mother had well-defined lymphatics, she also had areas with dermal backflow as shown in the inset images of (b)
Family evaluation
In addition to the proband and her mother, DNA was collected from 9 family members (subjects 146-154) including 5 members diagnosed with LE upon clinical examination. The pedigree chart in Fig. 2 shows that the diagnosed disorder segregates in an autosomal dominant manner. The affected members exhibited grade II bilateral leg lymphedema, with the exception of subject 154, with grade I bilateral leg lymphedema. DNA was extracted from blood samples using Paxgene Blood DNA Kit (PreAnalytix, Switzerland) according to the vendor's instructions. 2 μg of genomic DNA was submitted to Axeq Technologies for human exome capture sequencing using SureSelect Human All Exon V5 and V5 + UTRs (Agilent Technology, Santa Clara, CA).
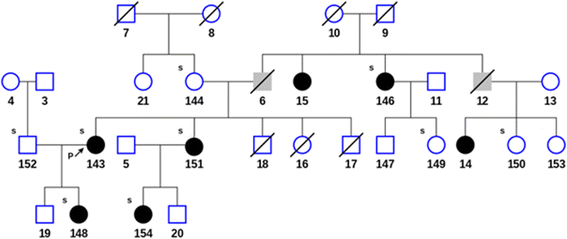
Fig. 2
Fig. 2 caption
Pedigree of the family showing members diagnosed with lymphedema (filled black), unknown phenotype (filled gray) and unaffected (open) subjects. Arrow indicates the proband. The small ‘s’ at the left top corner of the subjects indicates when the subject’s DNA was sequenced. A diagonal line indicates that the patient is deceased
Genetic sequencing analyses
Genetic sequencing analysis was conducted as follows. Each pair of fastq files was aligned to human genome (hg19) using Novoalign (Novocraft Technologies; www.novocraft.com), keeping parameters at the default settings, as recommended by Novocraft Technologies. SAMtools (http://samtools.sourceforge.net) was used to sort the SAM files, create BAM files, and generate their index files. Picard (SourceForge; http://broadinstitute.github.io/picard) was used to remove all of the PCR duplicates from the BAM files. For local realignments, base quality recalibration, and variant calling, we used Genome Analysis Toolkit (GATK) Version 3.1-1 [8]. Finally, for variant annotation, we used SnpEff (http://snpeff.sourceforge.net/) [9], variant tools and ANNOVAR [10] using multiple databases from UCSC Genome bioinformatics. Functional effects for each non-synonymous coding variant was evaluated using three different functional prediction algorithms: (1) Polyphen 2.0 Prediction of functional effects of human nsSNPs (http://genetics.bwh.harvard.edu/pph2) [11], (2) SIFT [12] and (3) (www.mutationtaster.org) [13] using the dbNSFP database [14]. Filtration of common polymorphisms was accomplished using frequencies from the NHLBI Exome sequencing project (ESP) (http://evs.gs.washington.edu/EVS) [15] and the 1,000 Genomes Project (http://www.1000genomes.org/data) [16].
WES results showed that the proband did not have variants in any of the known genes previously associated with lymphatic dysfunction. To maximize the power of the segregation analysis, we selected family members diagnosed with LE from each generation for a total of 5 affected cases. Since we did not have controls of the same ethnicity, we selected 4 unaffected family members to reduce the background and increase the statistical power of the analysis. The raw results from the sequencing passed our standard quality control, and the variant files (vcf) reported a consistent number of variants. After selecting non-synonymous coding variants with low frequency (MAF < 0.1), we performed co-segregation analysis using a dominant model of inheritance. Our analysis generated six potential candidate variants, five of them were eliminated after a quick manual inspection with the Integrative genomics viewer [17]. We identified a nonsense coding variant that generated a prematurely terminated protein CELSR1 (p.W1957X, Hg19chr22:g.46,790,132C > T; NC_000022.10:g.467901432C > T; NM_014246.1:c.5871G > A). The variant was private to the family and has not been reported in any of the following databases: 1000 Genomes [16], ESP6500 [15],UK10K, TCGA germline [18], Scripps Wellderly [19], ExAC [20], dbNSFP [14], ClinVar [21], OMIM [22], COSMIC [23], and Reference Variant Store (RVS), [24]. The variant sequence was of high quality with an average of 108.54 ± 20.93 reads per sample, mapping quality > 70, and Phred quality scores > 35. Furthermore, the variant was confirmed in each sample by Sanger sequencing in the forward direction using primer TGAGGTTGGGAGCCGGTAGAGG and the reverse direction using primer GTACCGACAGGGTATGTGAAGGCG (see Additional file 3: Figure S1). The segregation results were perfect, the variant was present in all the affected and was not present in any of the unaffected. We genotyped two additional unaffected members of the family, and they also lacked the variant. We searched using blat a 90 bp region containing the mutation against the human reference hg19 and found that the region was 100 % unique in the human genome. We also found that the same region was totally free of any repeats and evolutionarily conserved in vertebrates according to the comparative genomics track from the UCSC genome browser [25]. In summary, the ultra-rare damaging variant was present in five affected members and not present in six unaffected family members (Fig. 3a). From DNA of the family members sampled, there appears to be 100 % penetrance with an autosomal dominant pattern of inheritance.
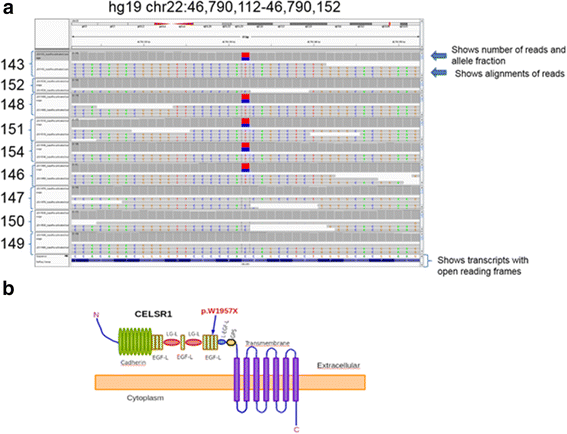
Fig. 3
Fig. 3 caption
a Image from the Integrative genomics viewer, showing a 40 bp genomic region containing the variant. Each one of the samples is loaded into the panel. Each panel shows two windows, the top window displays the number of reads, and the allele fraction and the lower window show the alignments of the experimental reads. The bottom panel displays the transcripts with the open reading frames. b A cartoon representing the predicted protein structure of CELSR1 (uniprot id Q9NYQ6). An arrow represents the location of the mutation. The cartoon was inspired by the publication of Lei, et al. 2014 []
Discussion
CELSR1 is a member of the cadherin superfamily, conserved during evolution with orthologs in invertebrate and vertebrates [26]. In addition, CELSR1 is an atypical cadherin involved in planar cell polarity. This protein has nine cadherin repeats, eight EGF-like domains, and seven transmembrane segments (Uniprot_id Q9NYQ6). CELSR1 is located at the plasma membrane with the cadherin domains acting as homophilic binding regions and the EGF-like domains involved in cell adhesion and receptor-ligand interactions. We postulate that when the mutated allele is expressed, the protein lacks the entire cadherin domain, the five EGF domains, and the LamG domains (Fig. 3b).
Recent biological evidence reported in the literature supports the causal role of
The proband displayed a common lymphatic phenotype consisting of extensive dermal backflow, tortuous vessels, as well as unique “sheet-like flow” in both legs indicating a defect in the valvular mechanism of lymph propulsion in collecting vessels. While we have not conducted functional studies of the
In this family, the phenotype was not apparent in any living males, but obviously was passed to daughters through male parents, both now deceased and not available for clinical diagnosis or DNA collection. Three living males in the third and fourth generations were not reported to have lymphedema, but unfortunately not available for clinical diagnosis and genotyping. As a result, we are unable to determine whether penetrance is limited to females.
Conclusion
This report provides the first evidence that defective planar cell polarity signaling pathway may participate as the cause of primary, non-syndromic lymphedema in humans.
Consent
Written informed consent for images contained in this report was obtained from the patient's parents in accordance with the guidelines of the University of Texas Health Science Center Institutional Review Board.
Sources of funding
Funded by the National Institutes of Health grants R01 HL092923 and U54 CA136404 (EMS-M).
Additional files
The abnormal NIRFLI movie of lymphatic function in the left foot of the proband showing unusual lymph flow from the dorsal foot injections. (AVI 4088 kb)
The normal NIRFLI movie of lymphatic function in the left foot of the asymptomatic mother showing typical lymph flow from the dorsal foot injections within collecting lymphatic vessels. (AVI 5480 kb)
A composite picture of two electropherograms comparing the Sanger sequencing results from two members of a family with lymphedema. The top panel represents the WT sequence of an unaffected family member (#152). The low panel represents the heterozygous mutation in the affected index patient (#143). The validation was performed as described previously [32] using the following primers: forward TGAGGTTGGGAGCCGGTAGAGG and reverse GTACCGACAGGGTATGTGAAGGCG. (TIF 201 kb)
References
- New developments in clinical aspects of lymphatic disease. J Clin Invest. 2014;124(3):915-921.
- Hereditary lymphedema: evidence for linkage and genetic heterogeneity. Hum Mol Genet. 1998;7(13):2073-2078.
- Genetics of lymphatic anomalies. J Clin Invest. 2014;124(3):898-904.
- Mutations in the VEGFR3 signaling pathway explain 36 % of familial lymphedema. Mol Syndromol. 2013;4(6):257-266.
- Translation of near-infrared fluorescence imaging technologies: emerging clinical applications. Annu Rev Med. 2012;63:217-231.
- Human Lymphatic Architecture and Dynamic Transport Imaged Using Near-infrared Fluorescence. Transl Oncol. 2010;3(6):362-372.
- Lymphatic imaging in humans with near-infrared fluorescence. Curr Opin Biotechnol. 2009;20(1):74-82.
- The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297-1303.
- A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6(2):80-92.
- Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10(10):1556-1566.
- Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;Chapter 7:Unit7 20.
- SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812-3814.
- MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7(8):575-576.
- Liu X, Wu C, Li C, Boerwinkle E. dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Non-synonymous and Splice Site SNVs. Hum Mutat. 2015. doi: 10.1002/humu.22932
- Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337(6090):64-69.
- A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061-1073.
- Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24-26.
- The Cancer Genome Atlas [http://cancergenome.nih.gov/].
- The Scripps Wellderly Study [ftp://stsi-ftp.sdsc.edu/pub/wellderly/].
- Exome Aggregation Consortium (ExAC) [http://exac.broadinstitute.org].
- ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862-868.
- OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43(Database issue):D789-798.
- COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue):D805-811.
- Integrating 400 million variants from 80,000 human samples with extensive annotations: towards a knowledge base to analyze disease cohorts. BMC Bioinformatics. 2016;17(1):24-.
- The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015;43(Database issue):D670-681.
- Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 2006;20(23):3199-3214.
- Potential dual molecular interaction of the Drosophila 7-pass transmembrane cadherin Flamingo in dendritic morphogenesis. J Cell Sci. 2006;119(Pt 6):1118-1129.
- The flamingo-related mouse Celsr family (Celsr1-3) genes exhibit distinct patterns of expression during embryonic development. Mech Dev. 2001;109(1):91-94.
- Role of the planar cell polarity gene CELSR1 in neural tube defects and caudal agenesis. Birth Defects Res A Clin Mol Teratol. 2012;94(3):176-181.
- Identification of novel CELSR1 mutations in spina bifida. PLoS One. 2014.9(3)
- Planar cell polarity protein Celsr1 regulates endothelial adherens junctions and directed cell rearrangements during valve morphogenesis. Dev Cell. 2013;26(1):31-44.
- Evidence for SH2 domain-containing 5'-inositol phosphatase-2 (SHIP2) contributing to a lymphatic dysfunction. PLoS One. 2014.9(11)