A brief primer on microRNAs and their roles in angiogenesis
Vascular Cell. 2013;
Received: 15 November 2012 | Accepted: 11 January 2013 | Published: 16 January 2013
Vascular Cell ISSN: 2045-824X
Abstract
Development of the vasculature is a complex, dynamic process orchestrated by a balance of pro and anti-angiogenic signaling pathways. The same signaling pathways are mis-regulated and exploited during pathological angiogenesis in cancer, inflammation and cardiovascular diseases and contribute to disease progression. In the last decade, small non-coding RNA molecules termed microRNAs (miRs) have emerged as key regulators of several cellular processes including angiogenesis. It is becoming clear that miRs function in complex networks and regulate gene expression both at the mRNA and protein levels thereby altering cellular signaling responses to specific stimuli. In the vasculature, miRs can function either in a pro-angiogenic manner and potentiate angiogenesis or act as anti-angiogenic miRs by enhancing cell death and decreasing endothelial proliferation. This review aims to provide an update on how microRNAs regulate gene expression and illustrate miR function in the vasculature with a discussion of potential applications of miRs as anti-angiogenic therapeutics.
Keywords
Angiogenesis microRNA networks miR-126 miR-132 p120RasGAP NOTCH Ocular angiogenesisHow are microRNAs made?
MicroRNAs (miRs) are small 20-24 nucleotide long RNAs that regulate gene expression by binding to mRNAs. miRs represent about 1-2% of the genes in several species including mammals [8]. Current version of miRbase reports 1600 precursors and 2042 mature miR sequences for humans and 855 precursors and 1281 mature miRs for mice [9]. It is estimated that miRs could well regulate 30-50% of protein coding genes. A large portion of miRs is encoded in introns of genes or in intergenic regions. They are transcribed as a primary miR (pri-miRNA) transcript by RNA polymerase II in conjunction with specific transcription factors that bind to a region upstream similar to a protein coding gene. These miR transcripts are then cleaved by a heterodimer of dsRNA binding proteins, Drosha and DiGeorge Syndrome Critical Region 8 (DGCR8), releasing a 55-70 nt small RNA hairpin termed pre-miR. This RNA hairpin is then exported out of the nucleus by Exportin-5 and Ran-GTP. In the cytoplasm, another RNAse III enzyme, Dicer cleaves the pre-miRs to generate 22nt duplex RNAs. These are now mature miRs that get incorporated into RNA induced silencing complex (termed RISC) consisting of two major classes of proteins Argonaute and GW182. While this is the canonical pathway of miR biogenesis, recent research has shown several alternative pathways depending on cell type, organism and biological contexts [10, 11]. For example, the loss of Dicer leads to a more significant phenotype in the brain and embryonic stem cells compared to the loss of either Drosha or DGCR8. Similarly, loss of Dicer in the vasculature has a more profound effect than the loss of Drosha suggesting there may be Drosha independent mechanisms of miR biogenesis [12]. These observations highlight the degree of complexity in the different RNA processing pathways.
How do miRs work?
When miRs were initially described, it was proposed that miRs bind to target mRNAs using a seed sequence consisting of 6-8 continuous bases and a few more complementary bases along the miR sequence. In plants, miRs have almost perfect complementarity with the target mRNA and these targets undergo direct RNAse mediated cleavage [13]. However, the mammalian miRs rarely had perfect complementarity with their targets. Therefore, it was hypothesized that mammalian miRs suppressed translation of target mRNAs leading to a decrease in target proteins [14]. Interestingly, recent work based on mRNA arrays and ribosome profiling has shown that miRs can also cause mRNA decay in mammalian cells and this is likely due to deadenylation of the mRNA [15]. The relative contributions of mRNA decay and translation inhibition mechanisms in suppressing the target protein levels remains an area of active investigation [16].
What do miRs do that proteins cannot do?
It has been postulated that miRs maintain robustness of gene expression by insulating biological systems against noise [17]. For example, miR-1 and miR-7 in Drosophila insulate muscle differentiation and photoreceptor determination from external perturbations [18, 19]. While there are other regulatory mechanisms that control gene expression programs such as transcriptional regulators, epigenetic modifications etc, miRs offer a unique advantage by regulating gene expression in the cytoplasm. Moreover, there are miRs that could be upregulated rapidly in cells up to several thousand copies per cell thereby achieving greater potency than otherwise possible. There have been simulations that demonstrate that miR mediated feed forward loops are more effective than transcriptional repressors in buffering gene expression against external perturbations [20]. Finally, the fact that miRs can regulate multiple targets, often several of them in a single pathway, makes them a more effective tool to modulate gene expression in response to specific stimuli.
Are miRs essential in the vasculature?
The importance of miRs in vascular development was apparent based on loss-of-function studies of Dicer. Dicer hypomorphic mice had severe vascular deformation in both the embryo and the yolk sac and died at E12.5-E-14.5 [21]. Similarly, loss of Dicer led to pericardial edema and impaired circulation in zebrafish embryos [22]. Moreover, endothelial specific deletion of Dicer in mice decreased postnatal angiogenesis in response to multiple stimuli [23]. Similarly, inducible deletion of Dicer in vascular smooth muscle cells in mice led to impaired contractility, smooth muscle differentiation and vascular remodeling [24]. Taken together, these early studies established that the loss of Dicer and the concomitant loss of miR processing activity caused significant structural and functional defects in the vascular compartment.
Which miRs regulate vascular function?
The first study to report a putative role for specific microRNAs in endothelial cells was performed by Poliseno et al. The authors used microarray based profiling and identified the most abundant miRs in HUVECs [25]. They reported that several miRs were predicted to target angiogenic growth factor receptors and characterized miR-221/222 as negative regulators of angiogenesis. One of the earliest miRs reported to affect endothelial function
While the majority of miRs that have been reported function as pro-angiogenic miRs, miR-17 ~ 92 cluster, a well-characterized group of miRs plays a critical role as a negative regulator of angiogenesis [30, 31]. Bonauer et al showed that miR-92a has an anti-angiogenic effect partly through targeting the integrin subunit α5. Loss of miR-92a improved functional recovery after myocardial infarction and limb ischemia by enhancing blood vessel growth [30, 31].
The miRs highlighted here are examples to illustrate the diverse ability of miRs in manipulating endothelial function. We and others have reviewed the many different miRs that play a role in vascular function [32, 33]. Some of the prominent miRs in endothelial cells and smooth muscle cells, their functions and target genes are listed in the table below (Table 1).
Table 1
| microRNA | Role | Target(s) | References |
|---|---|---|---|
| Let-7 family | Increases EC proliferation, migration | TIMP-1 | [ |
| miR-126 | Enhances VEGF signaling in ECs | SPRED1, PIK3R2, VCAM-1 | [ |
| miR-132 | Enhances growth factor signaling | p120RasGAP, PTEN | [ |
| miR-210 | Stimulates tube formation and migration | EphrinA3, NPTX1 | [ |
| miR-221/222 | Inhibits cell proliferation, Anti-angiogenic | C-kit, p27 | [ |
| miR-296 | Inhibits growth factor receptor recycling and degradation | Hepatocyte-growth factor regulated tyrosine kinase substrate | [ |
| miR-424 | Stablizes HIF-1, proangiogenic | Cullin-2 | [ |
| miR-92a | Anti-angiogenic. Inhibition enhanced neovascularization in ischemic tissues | ITGA5 | [ |
| miR-143/145 | Critical for VSMC differentiation | KLF4/KLF5 | [ |
| miR-26a | Increases VSMC proliferation | Smad-1/4 | [ |
miR-132 in endothelial and perivascular cells
We identified miR-132 as an angiogenic miR that functions by regulating p120RasGAP and Ras signaling pathways using a human embryonic stem cell vasculogenesis model and primary endothelial cells [36]. We demonstrated that reciprocal expression of miR-132 and its target p120RasGAP correlated with pathological angiogenesis in human breast tumors and in human hemangiomas. Targeted delivery of an anti-miR-132 decreased angiogenesis and tumor progression in mouse tumor xenograft models. Consistent with our observations in vascular endothelial cells, Lagos
Subsequent to our work, other groups have reported the function of miR-132 in perivascular cells. Natarajan and colleagues showed time- and dose-dependent up-regulation of miR-132/212 by Ang II through the Ang II Type 1 receptor [37]. They identified phosphatase and tensin homolog (PTEN) as a novel target of miR-132 and demonstrated that miR-132 induces monocyte chemoattractant protein-1 in rat VSMC. The authors demonstrated the presence of a positive feedback loop where phosphorylation of cAMP Response Element Binding protein (CREB) led to miR-132 transcription; miR-132 overexpression resulted in enhanced CREB phosphorylation via p120RasGAP downregulation. In addition, the authors observed that aortas from Ang II-infused mice displayed similar up-regulation of miR-132/212 and monocyte chemoattractant protein-1 and concluded that miR132/212 can serve as a novel cellular node to fine-tune and amplify Ang II actions in VSMC.
Another recent study [46] showed that miR-132 was constitutively expressed and secreted by saphenous vein pericytes and facilitates cardiac regeneration following myocardial infarction. Both CREB and miR-132 were activated by treatment of these cells by VEGF-B or hypoxia/starvation. The authors report that miR-132 is secreted by these pericytes and can act as a paracrine activator of cardiac healing, highlighting a non-cell autonomous role for miR-132.
What triggers miR expression?
The vasculature is exposed to several stimuli and stressors during developmental and pathological angiogenesis that trigger distinct miR expression profiles. For instance, miR-210 and miR-424 have been shown to be upregulated in response to hypoxia [38, 41]. We have observed specific miRs upregulated in response to angiogenic growth factors VEGF and bFGF [36]. Other studies have also documented miRs regulated by Notch pathway signaling [47] and cytokines such as IL-3 [48]. These stimuli subsequently trigger miR expression via classic signal transduction pathways and transcription factors. For example, miR-132 is transcribed by CREB in multiple cell types including endothelial cells, vascular smooth muscle cells and neurons [36, 37, 49]. Harris et al have shown that miR-126 is transcribed by Ets family members Ets-1 and Ets-2 [50]. Interestingly, Nicoli et al demonstrated that blood flow induced mechanosensitive transcription factor Klf2a can also transcribe miR-126 during aortic arch development in Zebrafish [28]. While the factors that induce miR expression have been widely characterized, regulatory sequences such as promoters for specific miRs have not yet been well defined and remain an area of considerable interest.
How do miR-target networks regulate angiogenic signals?
While initial reports of miR function focused on one or two specific targets of miRs, it is becoming clear that the biology of miR-target interaction is much more complicated than simple RNA-RNA binding that is the basis of target prediction algorithms. Recent evidence suggests that evolution of conserved miR/target networks can exert powerful influence on biological functions [51]. Given these considerations, one can envision future work in understanding physiological roles of miRs will include systems level analysis of multiple miRs and targets.
For example, it is well established that VEGF and Notch signaling are key regulators of developmental angiogenesis. Mapping of a few receptors, ligands and transcription factors on this pathway using STRING database [52] shows how these molecules interact with each other. Our work identified several microRNAs that were regulated during growth factor signaling in endothelial cells and human ES cell vascular development. Some of these miRs from our screen have been predicted to target specific NOTCH or VEGF signaling components Figure 1. Interestingly, it appears that the miRs that were downregulated (blue, italicized) are predicted to target NOTCH or VEGF signaling components. This observation implies that downregulation of a few miRs can amplify a pro-angiogenic signaling network. Conversely, upregulation of miRs such as miR-132 and miR-126 can potentiate angiogenesis by targeting negative regulators of VEGF signaling such as p120RasGAP, SPRED1 etc. This network illustrates a cascade of miR transcription or decay events that can function in a positive feedback loop to amplify signals and sustain angiogenic responses Figure 1.
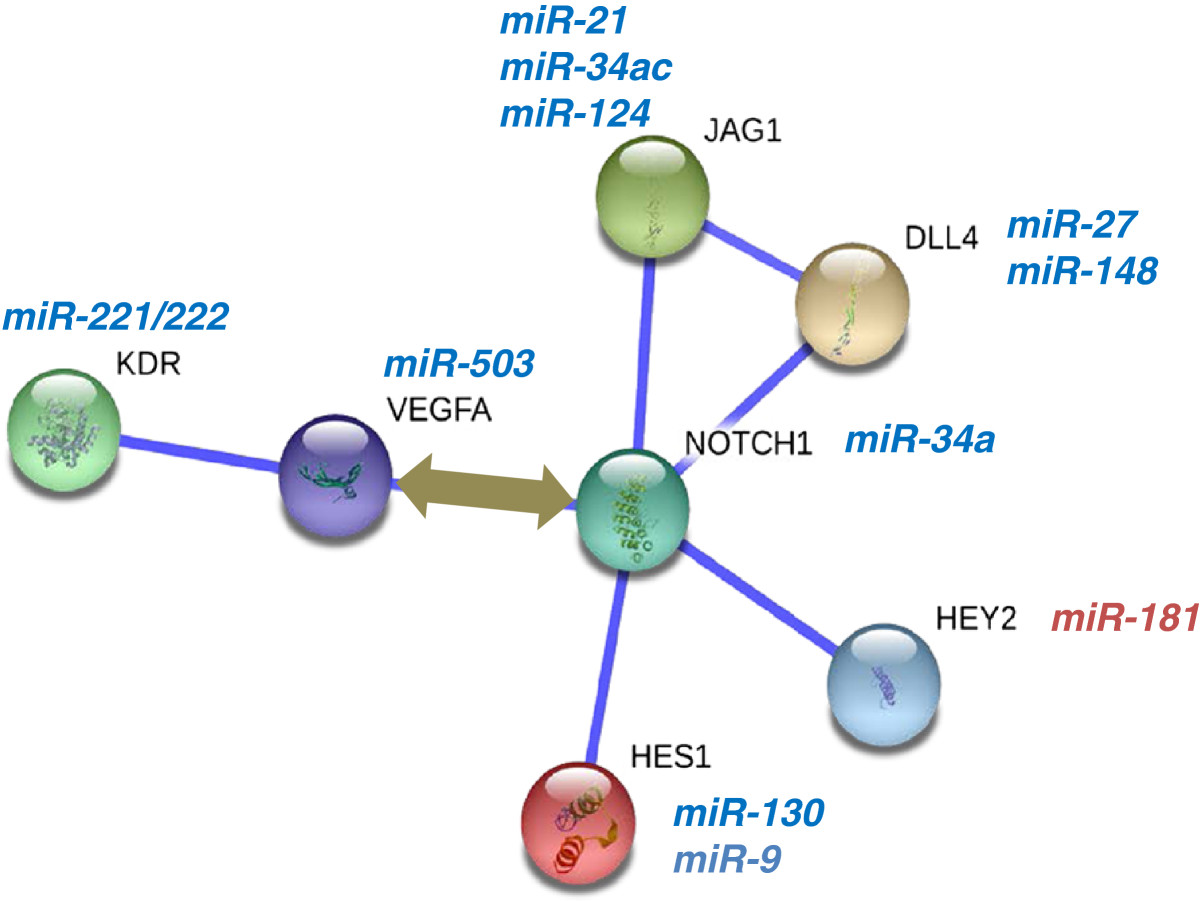
Figure 1
Figure 1 caption
A simple network of VEGF and Notch signaling regulated by microRNAs. miRs identified in angiogenic screens [] were cross-referenced with prediction programs for their ability to target specific nodes of a protein interaction network. Several miRs predicted to target angiogenic proteins were significantly downregulated in the screens (blue, italics). The illustration depicts how the loss of a few miRs that target positive regulators of angiogenesis can amplify a pro-angiogenic cascade.
Are miRs ‘druggable’ targets in human diseases?
Although miRs were discovered only about a decade ago, they have rapidly progressed to the verge of clinical utility. In fact, there are currently 124 clinical trials listed on microRNAs with 72 of them focused on cancer patients [53]. Many of these trials have focused on utilizing miR signatures for diagnostics across a spectrum of diseases. The first phase I trial of a microRNA therapeutic against HCV has shown excellent safety profile, good efficacy and has moved to Phase II trials highlighting that this novel class of drugs may have significant clinical potential [54]. Currently there are a number of different therapeutic strategies [55, 56] that are being evaluated in preclinical models to inhibit miRs as described in Table 2.
Table 2
Different strategies to inhibit miR function
| Reagent | Description |
|---|---|
| AntagomiRs | Cholesterol conjugated 2’-O-Methyl antisense oligonucleotides |
| LNA-AntimiRs | Locked nucleic acid based antisense oligos |
| miR sponges | Oligos with multiple miR binding sites to act as competitive inhibitors |
| miR-masks | Oligos with gene specific sequences complementary to miR binding sites |
Although these are exciting times for discovery of miRs and their applications, there are certain limitations to therapeutic targeting of miRs in the context of complex pathologies such as cancer.Targeting anti-miR therapeutics to specific tissues. Naked injection of anti-miRs or cholesterol conjugated antagomiRs typically results in excellent delivery to the liver. However, targeting specific anti-miRs to tissues such as tumor vasculature or sites of ischemic injury is still challenging. Nanoparticles and other delivery vehicles with targeting peptides may be useful in delivering anti-miRs for specific applications [55].The delivery of double stranded miR/miR mimics to tumor cells is still challenging and faces similar roadblocks as siRNA therapies. Recent work that shows that a single guide strand of RNA is sufficient to mediate RNA interference is exciting. This could lead to more efficient delivery of miRs to tumor cells
miR inhibitors as anti-angiogenic agents in ocular disease
miR inhibitors can be effective as simple oligonucleotides in ocular disease without the above mentioned requirements for specific delivery systems. Anti-angiogenic therapy, most notably, anti-VEGF antibody has shown significant therapeutic benefit in patients with Age-related Macular Degeneration (AMD) and consequently garnered several accolades including ‘
Interestingly, our work has identified several promising miRs that are downstream of VEGF signaling pathways that can be pursued as alternatives to VEGF inhibitors in the eye. We demonstrated that anti-miR-132 is able to decrease developmental angiogenesis in neonatal retinas without affecting established vasculature [36]. Since the intraocular injection of anti-miR reagents selectively interferes with angiogenic endothelial cells that have upregulated miR-132 and not the normal quiescent vasculature, it is likely that this strategy will not have the global effects observed with VEGF ablation. In collaboration with Friedlander and colleagues, we have now observed that anti-miR-132 is a potent inhibitor of pathological neovascularization in the eye in various mouse models (Westenskow and Friedlander, submitted). It remains to be seen if long term use of anti-miR-132 and more broadly anti-angiogenic miRs, can circumvent some of the adverse effects associated with the lack of VEGF in the eye while effectively inhibiting pathological neovascularization.
Conclusions
The emergence of miRs as regulators of gene expression has undoubtedly altered our understanding of how diverse stimuli affect function in the vasculature. miRs have been shown to alter specific signaling pathways that affect proliferation, differentiation, migration and cell survival in endothelial cells and smooth muscle cells. The challenges moving forward would be to integrate the different miRs and their target networks in the stimulus–response cycles of the vasculature and to understand how manipulating one or more miRs can alter cell function
Acknowledgements
S.A. is supported by a K99/R00 Pathway to Independence Award (HL112962) from NHLBI. I wish to acknowledge David Cheresh and members of the Cheresh Laboratory for useful discussions.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Authors’ original file for figure 1
References
- Endothelial cells are central orchestrators of cytokine amplification during influenza virus infection. Cell. 2011;146(6):980-991.
- Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481(7382):457-462.
- The vascular endothelium of the adipose tissue gives rise to both white and brown Fat cells. Cell Metabol. 2012;15(2):222-229.
- Developmental and pathological angiogenesis. Annual Rev Cell Develop Biol. 2011;27(1):563-584.
- Vascular microRNAs. Curr Drug Targets. 2010;11(8):943-949.
- MicroRNAs and vascular (dys)function. Vasc Pharmacol. 2011;55(4):92-105.
- Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469(7330):336-342.
- MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215-233.
- miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(suppl 1):D152-D157.
- Regulation of microRNA biogenesis and function. Thromb Haemost. 2012;107(4):605-610.
- Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Molecular cell. 2011;43(6):892-903.
- Role of dicer and drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101(1):59-68.
- Plant microRNA: a small regulatory molecule with big impact. Dev Biol. 2006;289(1):3-16.
- The impact of microRNAs on protein output. Nature. 2008;455(7209):64-71.
- Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835-840.
- Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12(2):99-110.
- Roles for MicroRNAs in conferring robustness to biological processes. Cell. 2012;149(3):515-524.
- A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137(2):273-282.
- Mesodermally expressed drosophila microRNA-1 is regulated by twist and is required in muscles during larval growth. Genes Dev. 2005;19(19):2343-2354.
- The role of incoherent MicroRNA-mediated feedforward loops in noise buffering. PLoS Comput Biol. 2011;7(3):e1001101-.
- Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;280(10):9330-9335.
- MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308(5723):833-838.
- Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci U S A. 2008;105(37):14082-14087.
- Smooth muscle miRNAs are critical for post-natal regulation of blood pressure and vascular function. PLoS One. 2011;6(4):e18869-.
- MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108(9):3068-3071.
- miR-126 regulates angiogenic signaling and vascular integrity. Develop cell. 2008;15(2):272-284.
- The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Develop cell. 2008;15(2):261-271.
- MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature. 2010;464(7292):1196-1200.
- A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2012;481(7380):190-194.
- MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324(5935):1710-1713.
- Members of the microRNA-17-92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood. 2010;115(23):4944-4950.
- Emerging role of micro-RNAs in the regulation of angiogenesis. Genes & cancer. 2011;2(12):1134-1138.
- MiRNAs as modulators of angiogenesis. Cold Spring Harbor perspect med. 2012.
- Impaired microRNA processing causes corpus luteum insufficiency and infertility in mice. J Clin Invest. 2008;118(5):1944-1954.
- MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci U S A. 2008;105(5):1516-1521.
- MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat med. 2010;16(8):909-914.
- Small RNA sequencing reveals MicroRNAs that modulate angiotensin II effects in vascular smooth muscle cells. J Biol Chem. 2012;287(19):15672-15683.
- MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283(23):15878-15883.
- Hypoxia induces microRNA miR-210 in vitro and in vivo ephrin-A3 and neuronal pentraxin 1 are potentially regulated by miR-210. FEBS Lett. 2008;582(16):2397-2401.
- miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer cell. 2008;14(5):382-393.
- Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-alpha isoforms and promotes angiogenesis. J Clin Invest. 2010;120(11):4141-4154.
- miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460(7256):705-710.
- MicroRNA-26a is a novel regulator of vascular smooth muscle cell function. J Cell Physiol. 2011;226(4):1035-1043.
- miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat cell biol. 2010;12(5):513-519.
- Role of miR-132 in angiogenesis after ocular infection with herpes simplex virus. Am J Pathol. 2012;181(2):525-534.
- Transplantation of human pericyte progenitor cells improves the repair of infarcted heart through activation of an angiogenic program involving micro-RNA-132/novelty and significance. Circ Res. 2011;109(8):894-906.
- The miR-143/145 cluster is a novel transcriptional target of Jagged-1/Notch signaling in vascular smooth muscle cells. J Biol Chem. 2011;286(32):28312-28321.
- microRNA-222 controls neovascularization by regulating signal transducer and activator of transcription 5A expression. Arterioscler Thromb Vasc Biol. 2010;30(8):1562-1568.
- An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci U S A. 2008;105(26):9093-9098.
- Ets-1 and Ets-2 regulate the expression of microRNA-126 in endothelial cells. Arterioscler Thromb Vasc Biol. 2010;30(10):1990-1997.
- Deep conservation of MicroRNA-target relationships and 3’UTR motifs in vertebrates, flies, and nematodes. Cold Spring Harbor Symposia Quantitative Biol. 2006;71:149-156.
- The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39(suppl 1):D561-D568.
- MicroRNA clinical trials from Clinical Trials.gov: http://clinicaltrials.gov/ct2/results?term=MICRORNA&Search=Search,
- Final Results - Randomized, Double-blind, Placebo-controlled Safety, Anti-viral Proof-of-Concept Study of Miravirsen, an Oligonucleotide Targeting miR-122, in Treatment-naïve Patients with Genotype1 (GT1) chronic HCV infection. In: 47th Annual Meeting of the European Association for the study of the liver: April 18-22. J Hepatol. 2012;56(suppl2):s26-.
- Targeting microRNAs in cancer: rationale, strategies and challenges. Nat rev Drug discov. 2010;9(10):775-789.
- MicroRNAs as potential cancer therapeutics. Oncogene. 2008;27(Suppl 2):S52-S57.
- Single-stranded siRNAs activate RNAi in animals. Cell. 2012;150(5):883-894.
- Single-stranded RNAs Use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell. 2012;150(5):895-908.
- Clinical application of therapies targeting VEGF. Cell. 2010;143(1):13-16.
- Pegaptanib for neovascular age-related macular degeneration. N Engl J Med. 2004;351(27):2805-2816.
- Targeted deletion of vegfa in adult mice induces vision loss. J Clin Invest. 2012;122(11):4213-4217.