Delta-like 4 mRNA is regulated by adjacent natural antisense transcripts
Vascular Cell. 2015;
Received: 19 November 2014 | Accepted: 11 March 2015 | Published: 24 March 2015
Vascular Cell ISSN: 2045-824X
Abstract
Background
Recent evidence suggests that a majority of RNAs in the genome do not code for proteins. They are located in the sense (S) or antisense (AS) orientation and, to date, the functional significance of these non-coding RNAs (ncRNAs) is poorly understood. Here, we examined the relationship between S and AS transcripts in the regulation of a key angiogenesis gene, Delta-like 4 (
Methods
Rapid Amplification of cDNA Ends (RACE) method was used to identify natural antisense transcripts in the
Results
Conclusion
A novel form of non-coding RNA-mediated regulation at the
Keywords
Non-coding RNA Delta-like4 Vascular HemangiomasBackground
Recent findings demonstrate that a new class of RNA arising from intergenic or introns of vascular specific genes participate in the regulation of angiogenesis, the growth of new blood vessels from existing vasculature [1-3]. These RNAs are referred to as non-coding RNAs (ncRNAs) because majority of ncRNAs in the genome do not code for proteins [4]. NcRNAs are classified as long (>200 bp) (lncRNAs) or short (<200 bp) (sncRNAs) based on their sizes [5]. Recent evidence suggests that they are located in the sense (S) or antisense (AS) orientation [6,7] and, to date, the functional significance of these ncRNAs is poorly understood. Previous work from our laboratory identified a non-coding RNA in the antisense direction to the
Methods
Identification of Dll4-AS
PolyA RNAs were isolated from mouse endothelial cell line, MS1 using a Poly(A)Purist Kit (Life Technologies, AM1916). FirstChoice RLM-RACE Kit (Life Technologies, AM1700) was used to obtain the full length sequences of
Subcellular localization of Dll4-AS
NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Scientific #78833) was used to separate nuclei and cytoplasm of MS1 cells. The separated two parts were subjected to TRIzol extraction, followed by reverse transcription.
Quantitative PCR
Total RNA from cultivated cells was extracted by TRIzol reagent followed by DNase I treatment for 2 h at 37°C. The DNA-free RNA was further purified using RNAeasy Mini kit (Qiagen, 74104). RNA concentrations were measured by Nanodrop (Thermo Scientific), followed by reverse transcription by SuperScript III (Life Sciences). Quantitative PCR was carried out with SYBR Green I in iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad, 170–9780). The expression levels were normalized to internal βactin or
Promoter reporter gene assay system
DNA fragments mapped to
Overexpression of Dll4-AS
pTracer™-CMV2 Vector (Life technologies, V885-01) was digested with EcoRV and NotI for sub-cloning
Dll4-AS knockdown
Vector-Based miRNA and synthetic siRNA were used to knockdown
Immunostaining
MS1 cells were plated on sterile glass coverslips placed in the wells of a culture plate and allowed to adhere overnight. Cells on coverslips were transfected with siRNAs. 48 h later, the coverslips were removed from the well, washed in TBS [50 mM Tris · HCl (pH 7.4), 150 mM NaCl], and fixed in 4% (wt/vol) paraformaldehyde for 10 min before permeabilization in 0.2% (vol/vol) Triton X-100 for 5 min. Cells were blocked in PBS with 1% goat serum and 0.1% Tween for 1 h at room temperature before staining with goat anti-mouse DLL4 (R&D, AF1389). Following 45 min incubation, cells were washed in TBS, followed by incubation with a fluorescent-conjugated IgG secondary antibody for 30 min in dark. After staining, coverslips were mounted in VectaShield containing DAPI.
Mouse studies
Care of the mice during experimental procedures was conducted in accordance with the policies of the Biomedical Resource Center, Medical College of Wisconsin, and the National Institutes of Health guidelines for the care and use of laboratory animals. Protocols had received prior approval from the Medical College of Wisconsin Institutional Animal Care and Use Committee. C57BL/6 mice were obtained from Charles River Laboratories (Franklin, CT). Six day-old C57BL/6 mouse pups were injected intraperitoneally with 1 mg/kg ultrapure LPS (Invivogen, CA) or saline and lungs were harvested after 18 h following sacrifice of animals. RNA was obtained from whole lung using the PureLink RNA kit from Life Technologies (Carlsbad, CA).
Cell proliferation assay
Cell proliferation was performed using an ELISA kit (Roche # 11647229001). 5 × 103 cells were inoculated into 96-well culture plate and cells were allowed to attach for 12 h. siRNA-lipid complexes containing 1 pmol mixed siRNA and 0.3 μl Lipofectamine RNAiMAX was added into each well. Medium was refreshed 48 h later and 10 μl BrdU labeling solution was added into it. 12 h later, the labeling medium was replaced by 200 μl FixDenat and incubated for 30 min at room temperature. The FixDenat was replaced by 100 μl anti-BrdU-POD working solution. 90 min later, the wells were washed 3 times with PBS. 100 μl substrate solution was added into each well. Photometric detection was performed 20 min later by SpectraMax 340PC384 Microplate Reader (Molecular Devices).
Cell migration assay
EOMA cells were plated onto a 6-well plate. After the cells adhered to the plate surface, control or mixed
Spheroid sprouting assay
MS1 cells in Dulbecco’s modified Eagle medium (DMEM) were suspended in hanging drops (300 cells/30 μL) on the underside of petridish lids. The hanging drops were incubated for 24 h to form spheroids. Harvested spheroids were suspended in 1.5% collagen gel, and the spheroids-containing collagen gel was rapidly transferred into 96-well plates pre-coated with the same collagen gel and allowed to polymerize at 37°C for 30 min. DMEM containing 30 ng/mL recombinant mouse VEGF was added to the plates to cultivate the cells for 7 days. Sprouting vessels were quantified under microscope by counting the sprouts that had grown out of each spheroid.
Results
Identification and characterization of Dll4-AS
We searched the mouse genome databases for non-coding RNAs at
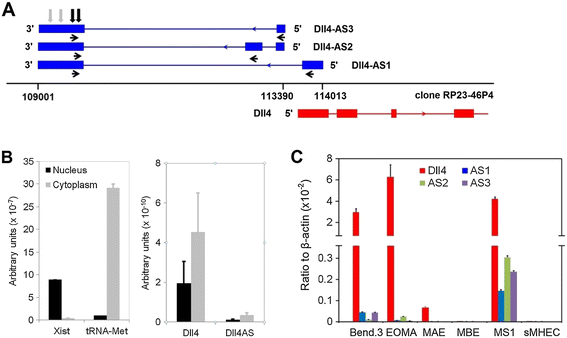
Figure 1
Figure 1 caption
Identification ofisoforms. (A) RACE reactions mapped the into promoter region. Blue and red arrowheads indicate transcriptional orientations of Dll4-AS and Dll4, respectively. Horizontal black arrowheads indicate following primers’ locations. Vertical black and grey arrows indicate following siRNA and miRNA targets, respectively. (B) is enriched in cellular cytoplasm. (C) expression coincides with in different cell lines. Bend.3: mouse endothelial cell from cerebral cortex; EOMA: mouse endothelial cell from hemangioendothelioma; MAE: Mouse Aortic Endothelial cell; MBE: Mouse Brain capillary Endothelial cell; MS1: mouse endothelial cell from pancreas; sMHEC: Mouse Heart Endothelial Cells.
Dll4-AS shares a common promoter with Dll4
To identify the genomic element that drives
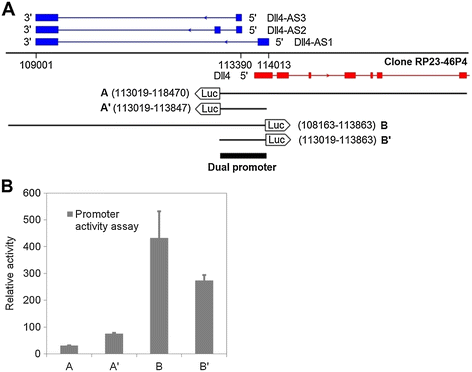
Figure 2
Figure 2 caption
Identification ofpromoter. (A) Genomic fragments aligning with both and transcription start sites were cloned into a promoter reporter vector, pGL4.14. (B) These constructed vectors were mixed with 1/20 (molar ratio) Renilla vector, which serves as internal control for cell transfection. The transfected MS1 cells were lysed 48 h later for luminescence reading.
Dll4-AS expression is concomitant with Dll4 mRNA expression
Because Notch-Delta signaling pathway has been extensively implicated in cell-cell contact signaling, we investigated the expression of
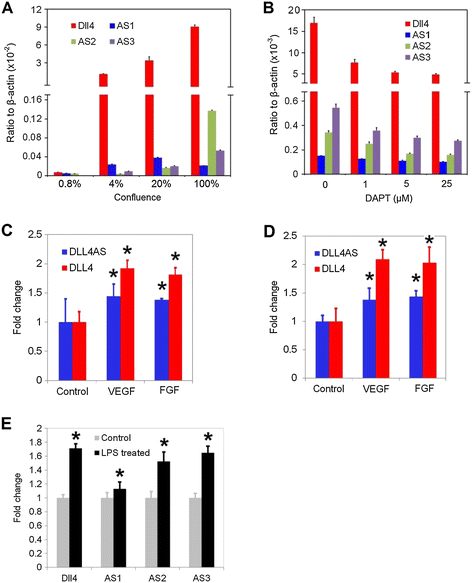
Figure 3
Figure 3 caption
responses to exogenous stimuli. (A) expression coincides with in the status of different cellular confluences. expression level was measured in 0.8%, 4%, 20% and 100% confluent MS1 cells by quantitative PCRs. (B) DAPT treatment down-regulates the expression of both and in MS1 cells. (C) HUVECs deprived of serum for 16 h were treated with 50 ng/ml recombinant human VEGF or FGF for 24 h. and were quantitated by qPCR in these cells. (D) HCAECs deprived of serum for 16 h were treated with 50 ng/ml recombinant human VEGF or FGF for 24 h. and were quantitated by qPCR in these cells. (E) 6-day old C57/BL6 mice were treated with 1 mg/kg intraperitoneal LPS. and were quantitated by qPCR in neonatal mouse lungs 18 hr after systemic LPS. *denotes < 0.05.
To confirm these findings in vivo, we performed experiments in the lung tissues in mice. Mice were treated with lipopolysaccharide (LPS) and lung tissues were harvested to analyze levels of
Dll4-AS regulates Dll4 expression
We investigated using loss-of-function approaches, the effect of
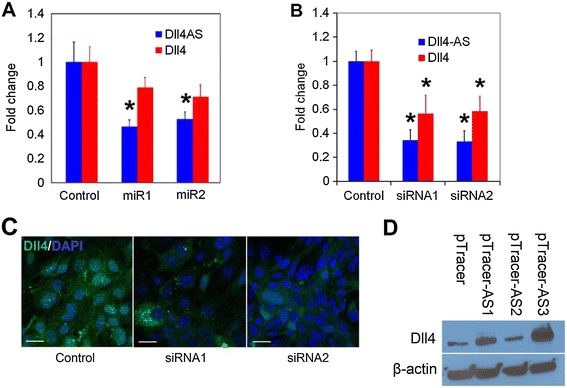
Figure 4
Figure 4 caption
Manipulation ofchanges the expression of. (A) pcDNA6.2-GW/EmGFP- miR vectors were transfected into MS1 cells. qPCR was performed 48 h later. Synthetic siRNAs were transfected into MS1 cells. qPCR (B) and immunostaining (C) were performed 48 h later. pTracer-Dll4-AS vectors were transfected into MS1 cells. Western blotting (D) was performed 48 h later.
Dll4-AS regulates angiogenesis
Because
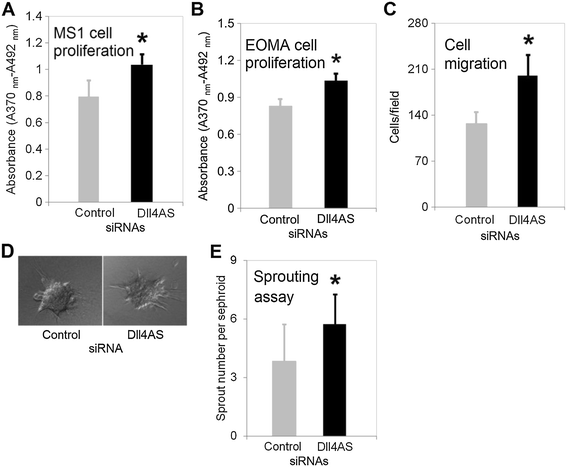
Figure 5
Figure 5 caption
Cell proliferation and migration assays. (A) MS1 cells in 96-well plate were transfected with 1 pmol synthetic siRNAs. 48 h later, transfection medium was replaced with regular medium containing BrdU labeling reagent. Incubation lasted for 16 h before proliferation assay was performed. *denotes < 0.05 (B) EOMA cells in 96-well plate were transfected with 1 pmol synthetic siRNAs. 48 h later, BrdU labeling solution was added into medium. Incubation lasted for 16 h before proliferation assay was performed. (C) Cell migration assay. EOMA cells in 6-well plate were transfected with 25 pmol synthetic siRNA. 48 h later, cells were placed onto trans-well membrane to allow migrate for 2 h. Cells were counted under microscope. (D) Representative spheroids with sprouting vessels. MS1 cells in 6-well plate were transfected with 25 pmol synthetic siRNA. 24 h later, cells were suspended in hanging drops to allow cellular aggregation. Spheroids were embedded in collagen gel to culture for 7 days. (E) Quantification of sprouts growing from the spheroids. 30 spheroids were examined for each transfection. *denotes < 0.05.
Discussion
In this study, we have identified lncRNAs at the
LncRNAs are a new class of regulators involved in genome organization and gene expression, especially in the process of cell differentiation and organ development [17]. In contrast to sncRNAs such as miRNAs, which target multiple coding sequences, lncRNAs usually target nearby genes [2,18]. Haploinsufficient genes like
Antisense ncRNAs arising from promoter regions can be classified into two categories according to their location [20]. The first category is composed of antisense ncRNAs overlapping with the corresponding mRNAs like
Genome locus of
Transcriptional correlation prognosticates functional association [27], so the function of
Conclusion
In summary, we report here the identification of three lncRNAs in the antisense direction in the
Acknowledgements
The authors thank members of the Developmental Vascular Biology Program for their critical input and suggestions, Dr. Auerbach at the University of Wisconsin-Madison for providing murine endothelial cell lines during the course of this work. This work was supported by a postdoctoral fellowship (K.L.) from American Heart Association ([10] POST4180008) and grants from National Institutes of Health [1R01HL102745-01 & 1R01HL112639-01] (R.R.). T.C. is supported by funds from Department of OBGYN. VS is partly supported by the CTSI of Southeast Wisconsin 8KL2TR000056 grants.
Additional file
Schematic representation of DLL4 locus in Homo sapiens chromosome 15, GRCh38 Primary Assembly. DLL4 mRNA starting from 40929333 is shown in red boxes; DLL4-AS starting from 40927667 and ending at 40926900 is shown in a blue box.
References
- A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood. 2010;115:133-9.
- Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114:1389-97.
- Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler Thromb Vasc Biol. 2014;34:1249-59.
- Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240-51.
- Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11-42.
- The antisense transcriptomes of human cells. Science. 2008;322:1855-7.
- Systematic identification of sense-antisense transcripts in mammalian cells. Nat Biotechnol. 2004;22:104-8.
- Endothelial cells and VEGF in vascular development. Nature. 2005;438:937-45.
- Haploinsufficiency of delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. Proc Natl Acad Sci U S A. 2004;101:15949-54.
- Dll4, a novel Notch ligand expressed in arterial endothelium. Genes Dev. 2000;14:1313-8.
- Expression of Dll4 during mouse embryogenesis suggests multiple developmental roles. Gene Expr Patterns. 2005;5:750-5.
- Bacterial lipopolysaccharide directly induces angiogenesis through TRAF6-mediated activation of NF-kappaB and c-Jun N-terminal kinase. Blood. 2003;102:1740-2.
- Downregulation by lipopolysaccharide of Notch signaling, via nitric oxide. J Cell Sci. 2008;121:1466-76.
- Endothelial cells dynamically compete for the tip cell position during angiogenic sprouting. Nat Cell Biol. 2010;12:943-53.
- Notch-1 signalling is activated in brain arteriovenous malformations in humans. Brain. 2009;132:11-.
- Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature. 2006;444:1032-7.
- The rise of regulatory RNA. Nat Rev Genet. 2014;15:423-37.
- Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494-504.
- Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells: implications for modulating arteriogenesis and angiogenesis. Mol Cell Biol. 2003;23:14-25.
- Bidirectional promoters are the major source of gene activation-associated non-coding RNAs in mammals. BMC Genomics. 2014;15:35-.
- PU.1 expression is modulated by the balance of functional sense and antisense RNAs regulated by a shared cis-regulatory element. Genes Dev. 2008;22:2085-92.
- Single-stranded noncoding RNAs mediate local epigenetic alterations at gene promoters in rat cell lines. J Biol Chem. 2011;286:34788-99.
- Analysis of Dll4 regulation reveals a combinatorial role for Sox and Notch in arterial development. Proc Natl Acad Sci U S A. 2013;110:11893-8.
- Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845-8.
- Differential effects of DNA-binding proteins on bidirectional transcription from the common promoter region of human collagen type IV genes COL4A1 and COL4A2. Biochim Biophys Acta. 1993;1174:1-10.
- An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62-6.
- Predicting gene function by conserved co-expression. Trends Genet. 2003;19:238-42.
- Systematic analysis of head-to-head gene organization: evolutionary conservation and potential biological relevance. PLoS Comput Biol. 2006;2:e74-.
- Deregulated expression of the RanBP1 gene alters cell cycle progression in murine fibroblasts. J Cell Sci. 1997;110(Pt 19):2345-57.
- Genomic structure of the human mitochondrial chaperonin genes: HSP60 and HSP10 are localised head to head on chromosome 2 separated by a bidirectional promoter. Hum Genet. 2003;112:71-7.
- Directional regulatory activity of cis-acting elements in the bidirectional alpha 1(IV) and alpha 2(IV) collagen gene promoter. J Biol Chem. 1993;268:24677-82.
- Notch signalling limits angiogenic cell behaviour in developing zebrafish arteries. Nature. 2007;445:781-4.